Our crossMoDA challenge at MICCAI 2021 is now live!
CAI4CAI members are leading the organization of the cross-modality Domain Adaptation challenge (crossMoDA) for medical image segmentation Challenge, which runs as an official challenge during the Medical Image Computing and Computer Assisted Interventions (MICCAI) 2021 conference.
Domain Adaptation (DA) has recently raised strong interests in the medical imaging community. By encouraging algorithms to be robust to unseen situations or different input data domains, Domain Adaptation improves the applicability of machine learning approaches to various clinical settings. While a large variety of DA techniques has been proposed for image segmentation, most of these techniques have been validated either on private datasets or on small publicly available datasets. Moreover, these datasets mostly address single-class problems.
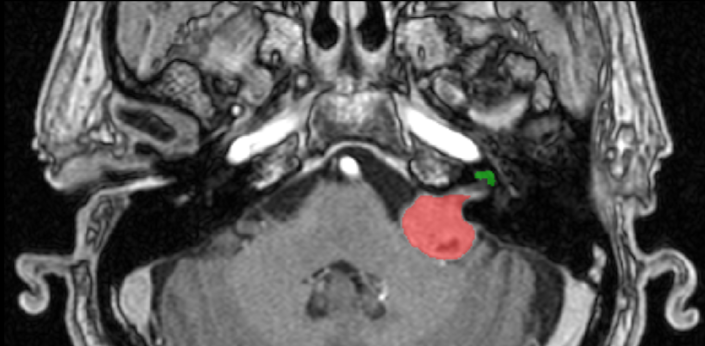
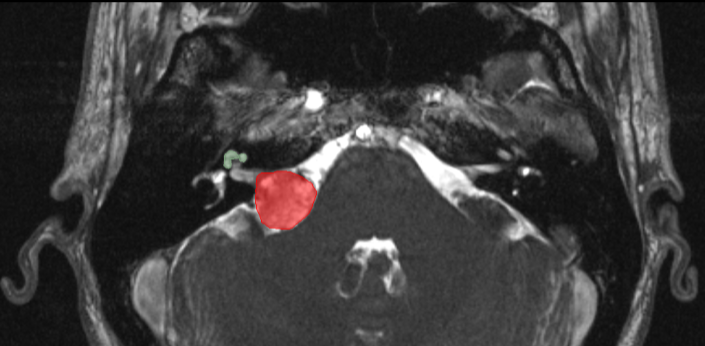
To tackle these limitations, the crossMoDA challenge introduces the first large and multi-class dataset for unsupervised cross-modality Domain Adaptation. The goal of the challenge is to segment two key brain structures involved in the follow-up and treatment planning of vestibular schwannoma (VS): the tumour and the cochlea. Specifically, the segmentation of the tumour and the surrounding organs at risk, such as the cochlea, is required for radiosurgery, a common VS treatment. Moreover, tumour volume measurement has also been shown to be the most accurate measurements for the evaluation of VS growth. While contrast-enhanced T1 (ceT1) Magnetic Resonance Imaging (MRI) scans are commonly used for VS segmentation, recent work has demonstrated that high-resolution T2 (hrT2) imaging could be a reliable, safer, and lower-cost alternative to ceT1. For these reasons, we propose an unsupervised cross-modality challenge (from ceT1 to hrT2) that aims to automatically perform VS and cochlea segmentation on hrT2 scans. The training source and target sets are respectively unpaired annotated ceT1 and non-annotated hrT2 scans.
This challenge will be the first medical segmentation benchmark of unsupervised DA techniques and promote the development of new unsupervised domain adaptation solutions for medical image segmentation. It will also contribute to the development of new algorithms for the follow-up and treatment planning of VS using hrT2 scans only.
More information about the challenge here.